Fragmentation and dissolution rates#
Fragmentation rate \(k_\text{frag}\) and dissolution rate \(k_\text{diss}\) control the rate at which polymer mass is lost from particle size classes, either to smaller size classes (fragmentation) or dissolved organics (dissolution). They have units of inverse time (s-1 in SI units) and thus, for a given period of time \(t + \Delta t\), a mass \(m = \Delta t k_{i,t}\) of polymer fragments or dissolves from size class \(i\). The Fragment size distribution controls how this mass is split amongst daughter size classes.
Both rate constants can vary in time and across particle surface areas, and thus they are 2D arrays with dimensions (n_timesteps, n_size_classes). This dependence is controlled by the parameters provided in model input data, specifically to the k_frag and k_diss variables. If scalars are provided to these, the rates are constant in time and surface area, and if a dict of regression parameters is provided, the rate constants vary in time and surface area according to these parameters, as detailed here.
The rate constant distributions are built using either a compound relationship:
or an additive relationship:
You can specify which is used by specifying the boolean is_compound in the k_frag or k_diss input data dict. Here, t is the time dimension, s is the particle surface area dimension, k_f is the rate constant scaling factor, k_0 is the rate constant baseline adjustment factor and k could be k_frag or k_diss. T(t) is the expression used to create the time-dependence distribution, and S(s) the surface area distribution. Both of these take the same form, which allows for power law, exponential, logarithmic and logistic regressions:
or if the user specifies a list for parameter \(A_x\), a polynomial instead of power law can be used:
Here, X(x) is either T(t) or S(s). The parameters A_t, A_s, alpha_t etc are the parameters provided to the k_frag and k_diss dicts in the input data. The dimension value \(\hat{x}\) is normalised such that the median value is equal to 1: \(\hat{x} = x/\tilde{x}\), where \(\tilde{x}\) is the median of \(x\). This ensures regressions are not skewed by t and s values that are many orders of magnitude apart.
Smallest size class
The model presumes that no fragmentation happens from the smallest size class, and so k_frag is always set to 0 for this size class. This is in constast to dissolution, which can happen from this size class.
Regression parameters#
Parameter |
Type, default |
Description |
|---|---|---|
|
Float, required |
Rate constant scaling factor |
|
Float, default=0 |
Rate constant baseline adjustment factor |
|
Float or list, default=1 |
Power law coefficient. If a scalar is provided, this is used as the coefficient for a power law expression with |
|
Float, default=0 |
If |
|
Float, default=1 |
Exponential coefficient |
|
Float, default=0 |
Exponential scaling factor |
|
Float or None, default=None |
If a scalar is given, this is the coefficient for the natural logarithmic term. If |
|
Float, default=1 |
Logarithmic scaling factor |
|
Float or None, default=None |
If a scalar is given, this is the coefficient for the logistic term. If |
|
Float, default=1 |
Logistic growth rate (steepness of the logistic term) |
|
Float or None, default=None |
Midpoint of the logistic curve, which denotes the |
|
Bool, default=True |
Whether the |
Negative k values
Make sure to specify regression parameters that do not end up in negative k values. The model checks for these before running and will throw a FMNPDistributionValueError exception if any are found.
Examples#
The make this more understandable, here are some examples are how you might use these parameters in real life. The first thing to note is that, if you want a k distribution to follow one particular regression, such as logistic, then you only need to specify parameters for that regression and the defaults take care of making sure the other regressions are constant. Taking a logistic regression as an example for k_frag:
import numpy as np
import matplotlib.pyplot as plt
from fragmentmnp import FragmentMNP
from fragmentmnp.examples import minimal_config, minimal_data
# By specifying D_t=1, we are telling the model that we want a
# logistic regression over time for k_frag. delta1_t controls
# the steepness of the logistic
minimal_data['k_frag'] = {
'k_f': 0.01,
'D_t': 1,
'delta1_t': 10
}
# Init the model with these data, which generates the k distributions
fmnp = FragmentMNP(minimal_config, minimal_data)
# Plot the k_frag distribution to check that it is logistic. We
# didn't specify any surface area dependence params and so we
# can pick any size class except the smallest (which always
# has k_frag=0)
plt.plot(fmnp.t_grid, fmnp.k_frag[1, :])
plt.xlabel('time')
plt.ylabel('k_frag')
Text(0, 0.5, 'k_frag')
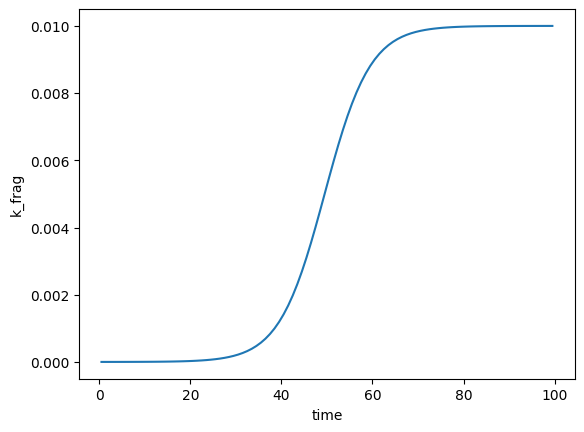
In the above, we effectively specified that k_frag should follow the regresion \(k_\text{frag} = 0.01 / (1 + e^{-(\hat{t} - 1)})\). Let’s now introduce a linear surface area dependence. Because the particle size classes are log-spaced, we plot with a log yscale in order to see the fragmentation rates for the smaller size classes:
# Specify a logistic regression over t and linear over s
minimal_data['k_frag'] = {
'k_f': 0.01,
'D_t': 1,
'delta1_t': 10,
'A_s': [0.01]
}
# Init the model
fmnp = FragmentMNP(minimal_config, minimal_data)
# Plot k_frag for all size classes except the smallest
plt.plot(fmnp.t_grid, fmnp.k_frag[1:, :].T)
plt.legend([f'{c * 1e6:<1g} µm' for c in fmnp.psd[1:]])
plt.xlabel('time')
plt.ylabel('k_frag')
plt.yscale('log')
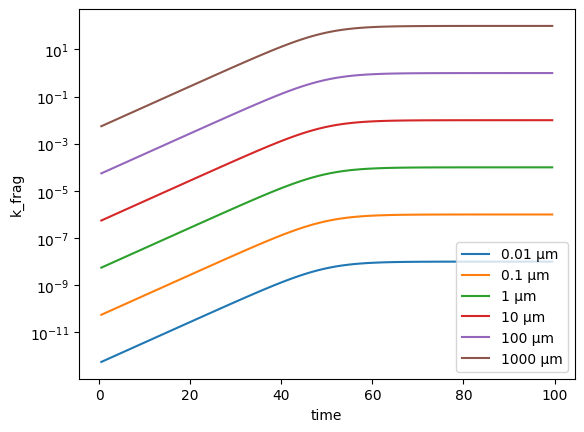
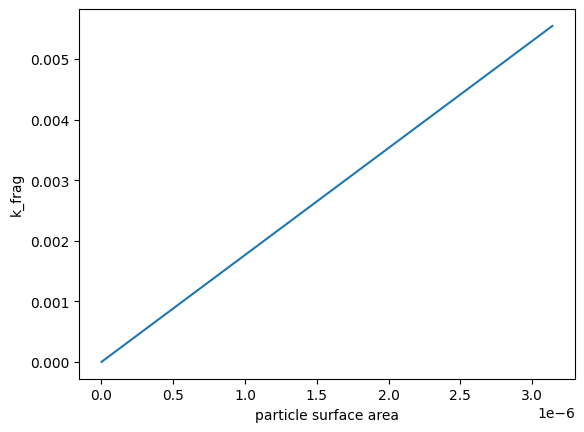
We can double check that the surface area dependence is linear by plotting the surface area against k_frag for one timestep:
plt.plot(fmnp.surface_areas[1:], fmnp.k_frag[1:, 0])
plt.xlabel('particle surface area')
plt.ylabel('k_frag')
Text(0, 0.5, 'k_frag')
Combining regressions can make for some weird and wonderful distribution shapes:
# Third-order polynomial combined with a logarithmic regression - why not!?
minimal_data['k_frag'] = {
'k_f': 0.01,
'k_0': 1,
'A_t': [-5, -2, 3],
'C_t': 1
}
# Create and plot
fmnp = FragmentMNP(minimal_config, minimal_data)
plt.plot(fmnp.t_grid, fmnp.k_frag[1, :])
plt.xlabel('time')
plt.ylabel('k_frag')
Text(0, 0.5, 'k_frag')
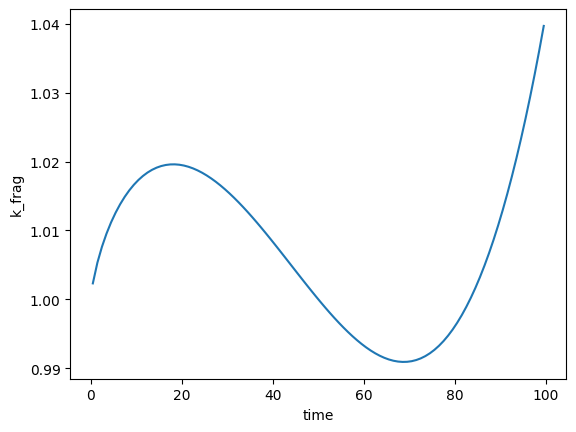
Setting your own \(k_\text{frag}\) array#
If the time and size dependencies that can be achieved using the regression parammeters above are not flexible enough for your scenario, you can of course set your own \(k_\text{frag}\) or \(k_\text{diss}\) arrays by modifying the k_frag or k_diss attributes directly after initialising the model:
# Set a random k_frag array (really?!)
fmnp.k_frag = np.random.rand(fmnp.n_size_classes, fmnp.n_timesteps)